Figure S7. Properties of genes with multiple causal variants.#
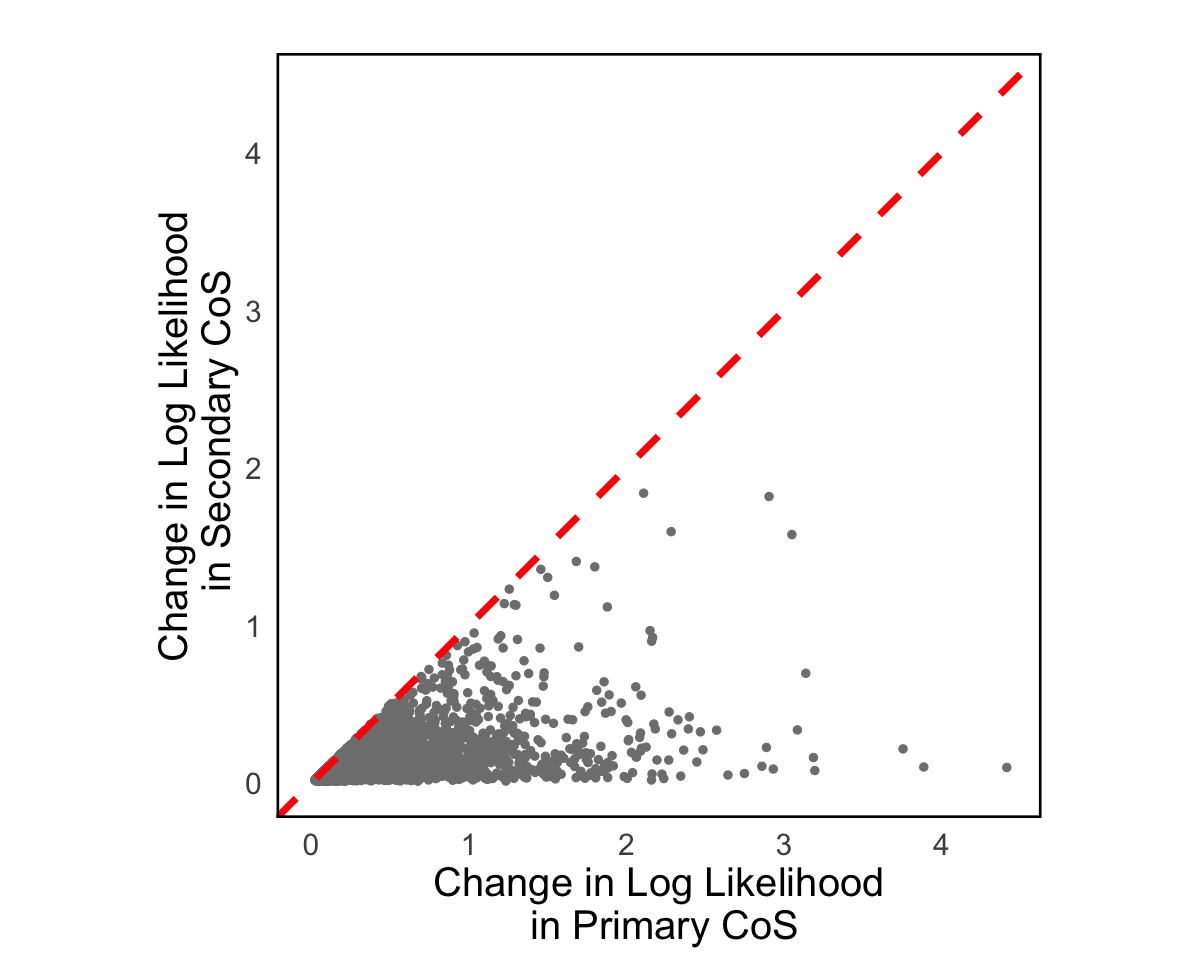
S7a. Scatter plot of the change in log-likelihood between primary CoS and a secondary CoS, where each point is a primary-secondary CoS pair for the gene.
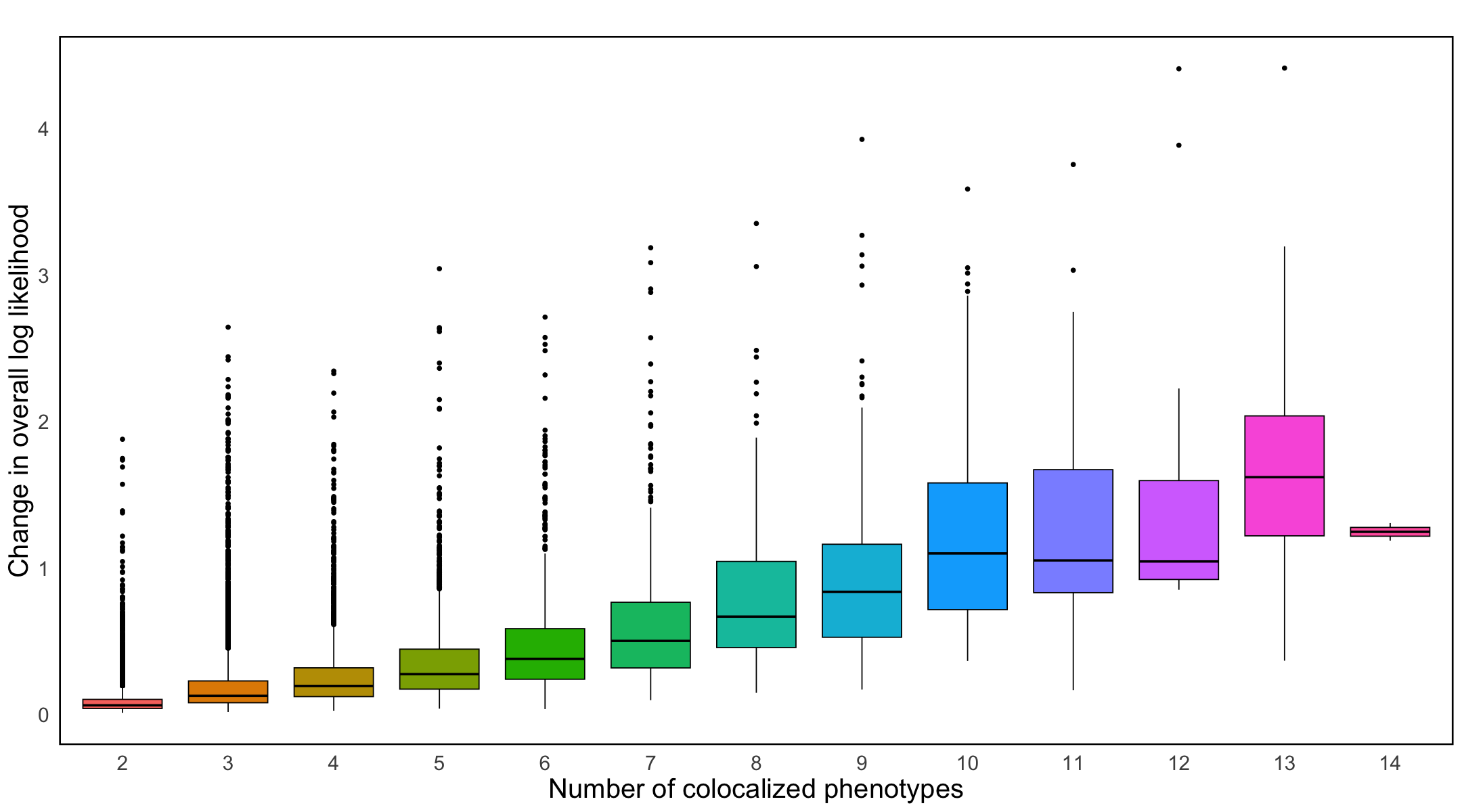
S7b. A boxplot summarizing the relationship between the number of colocalized traits and log likelihood changes.
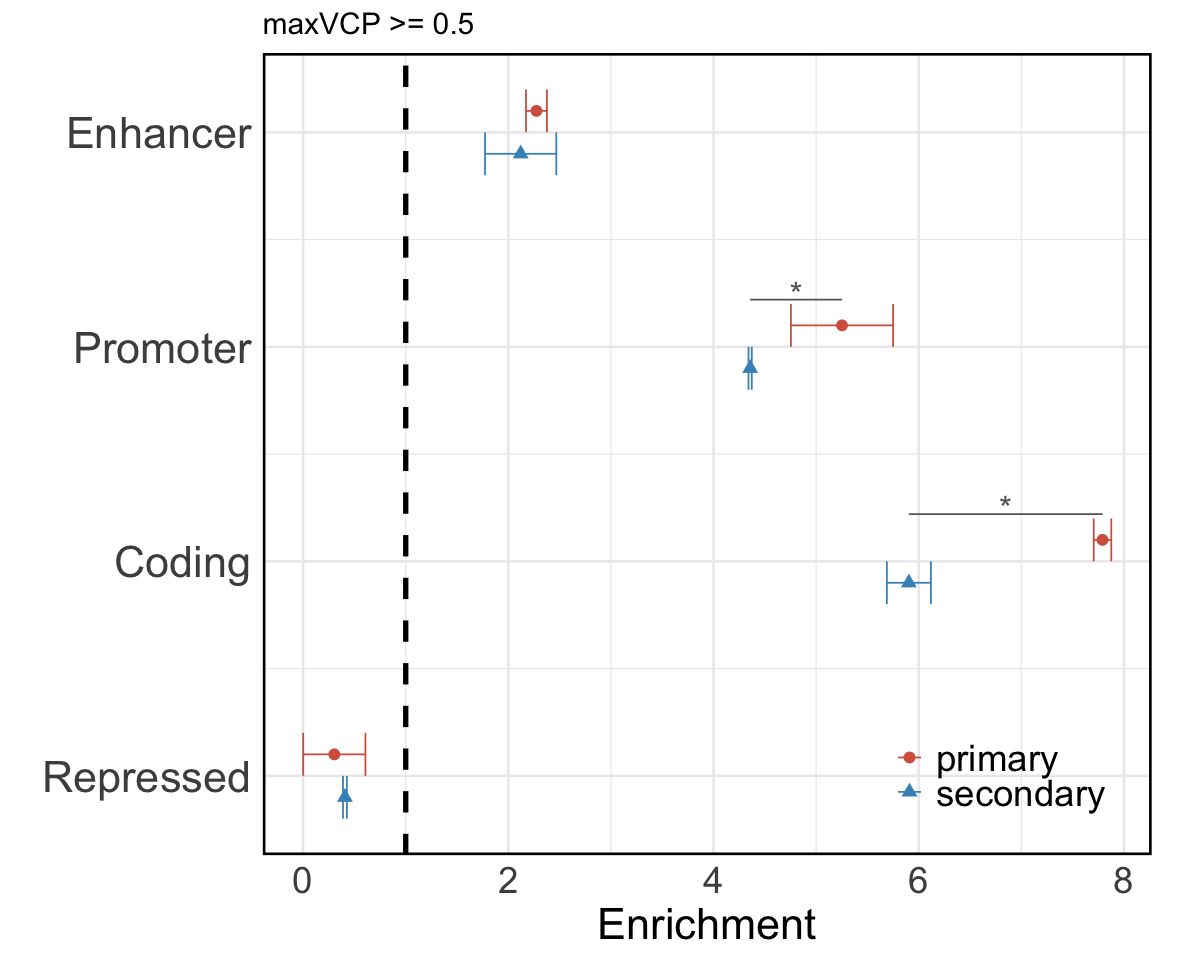
S7c. Functional annotation enrichment comparison between high MaxVCP scored variants (MaxVCP>0.5) in primary CoS versus secondary CoS, including annotations from enhancer, promoter, coding, and repressed categories.
S7d. An example case of xQTL-only colocalization in gene ARSB showing two CoS, one colocalized among all brain cell types and the other colocalized across microglia, astrocytes and oligodendrocytes only.
Figure S7a#
Scatter plot of the change in log-likelihood between primary CoS and a secondary CoS, where each point is a primary-secondary CoS pair for the gene.
result <- readRDS("Figure_S7a.rds")
dim_max <- max(c(result$primary_loglik, result$secondary_loglik))
p1 <- ggplot(result, aes(x = primary_loglik, y = secondary_loglik)) +
geom_point(size = 2, color = "grey50") +
geom_abline(slope = 1, intercept = 0, linetype = "dashed", color = "red", linewidth = 2) +
coord_fixed(ratio = 1, xlim = c(0, dim_max), ylim = c(0, dim_max)) +
labs(title = "",
x = "Change in Log Likelihood\n in Primary CoS",
y = "Change in Log Likelihood\n in Secondary CoS") +
theme_minimal(base_size = 15) +
theme(
axis.text.x = element_text(margin = margin(t = 5), size = 18), # Adjust x axis text margin
axis.text.y = element_text(margin = margin(r = 5), size = 18), # Adjust y axis text margin
axis.title.x = element_text(size = 24),
axis.title.y = element_text(size = 24),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_rect(color = "black", fill = NA, linewidth = 1.5))
Figure S7b#
A boxplot summarizing the relationship between the number of colocalized traits and log likelihood changes.
library(tidyverse)
res = readRDS("../../Main_Figures/Figure_3/data/xQTL_only_colocalization.rds")
coloc_pheno_numbers <- sapply(res$colocalized_phenotypes, function(cp){
length(strsplit(cp, "; ")[[1]])
})
res <- res %>% mutate(num_phen = coloc_pheno_numbers)
res <- within(res, {
num_phen <- factor(num_phen, levels = c(1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17))
})
library(ggsci)
color <- c(pal_npg()(10), pal_d3()(10))
p1 <- ggplot(res, aes(x = num_phen, y = profile_change, fill = num_phen)) +
geom_boxplot(color = "black") +
scale_color_manual(values = color) +
labs(title = "",
x = "Number of colocalized phenotypes",
y = "Change in overall log likelihood",
fill = "") +
theme_minimal(base_size = 15) +
theme(
axis.text.x = element_text(margin = margin(t = 5), size = 18), # Adjust x axis text margin
axis.text.y = element_text(margin = margin(r = 5), size = 18), # Adjust y axis text margin
axis.title.x = element_text(size = 24),
axis.title.y = element_text(size = 24),
legend.position = "",
panel.grid.minor = element_blank(),
panel.grid.major = element_blank(),
panel.border = element_rect(color = "black", fill = NA, linewidth = 1.5)
) +
guides(fill = guide_legend(nrow = 1))
Figure S7c#
Functional annotation enrichment comparison between high MaxVCP scored variants (MaxVCP>0.5) in primary CoS versus secondary CoS, including annotations from enhancer, promoter, coding, and repressed categories.
library(ggsci)
Enrichment_long = readRDS("Figure_S7c.rds")
p2 <- ggplot(Enrichment_long, aes(x = Enrichment, y = as.numeric(Category) + ifelse(Type == "primary", 0.1, -0.1), color = Type, shape = Type)) +
geom_point(size = 3) +
geom_errorbarh(aes(xmin = Enrichment - sd, xmax = Enrichment + sd), height = 0.2) +
scale_color_manual(values = c("primary" = "#D6604D", "secondary" = "#4393C3")) +
scale_y_continuous(breaks = 1:4, labels = levels(Enrichment_long$Category), sec.axis = dup_axis()) +
labs(
title = "maxVCP >= 0.5",
x = "Enrichment",
y = "",
color = "",
shape = ""
) +
theme_minimal(base_size = 15) +
theme(
axis.text.y.left = element_text(size = 26),
axis.text.y.right = element_blank(),
axis.title.x = element_text(size = 26),
axis.text.x = element_text(size = 22),
legend.text = element_text(size = 22),
panel.border = element_rect(color = "black", fill = NA, linewidth = 1.5),
legend.position = "inside",
legend.justification = c(0.95, 0.05)
) +
geom_vline(xintercept = 1, linetype = "dashed", linewidth = 1.5)
# Adding short lines and asterisks
for (cat in c("Promoter","Coding")) {
primary_point <- Enrichment_long[Enrichment_long$Category == cat & Enrichment_long$Type == "primary", ]
secondary_point <- Enrichment_long[Enrichment_long$Category == cat & Enrichment_long$Type == "secondary", ]
if (nrow(primary_point) == 1 && nrow(secondary_point) == 1) {
y_primary <- as.numeric(primary_point$Category)
y_secondary <- as.numeric(secondary_point$Category)
x_primary <- primary_point$Enrichment
x_secondary <- secondary_point$Enrichment
p2 <- p2 +
annotate("segment", x = x_primary, xend = x_secondary, y = y_primary + 0.22, yend = y_primary + 0.22, color = "grey40") +
annotate("text", x = mean(c(x_primary, x_secondary)), y = y_primary + 0.25, label = "*", size = 7, color = "grey40")
}
}