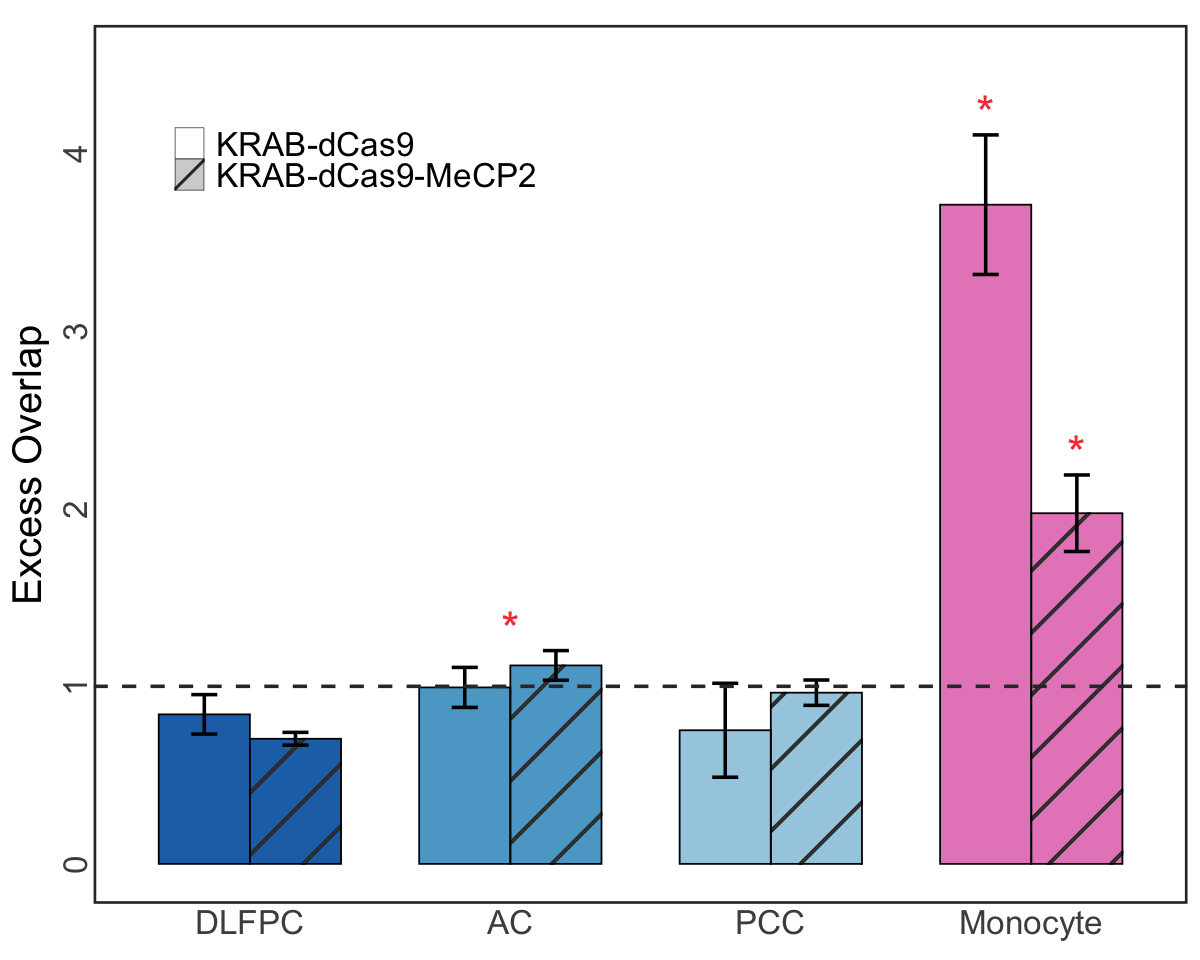
Figure 4b. Variant set -level Excess-of-overlap (EOO) analysis in bulk region.#
Variant set-level Excess-of-overlap (EOO) analysis of the 95% CoS-gene links across 3 different brain cortical regions and CD14+/CD16- bulk monocytes against the gold standard element-gene links from aggregated CRISPR (dCas9) and STING-seq datasets. Red asterisks denotes significant enrichment based on EOO test adjusted for multiple testing (FDR<5% using Bonferroni correction).
library(ggplot2)
library(ggsci)
library(ggpattern)
data <- readRDS("data/Figure_4b_bulk_EOO.rds")
sdtimes <- 1.96
colors <- c("#2171B5", "#5CA7CE", "#A6CEE3", "#E78AC3")
p1 <- ggplot(data, aes(x = celltype, y = enrichment, fill = celltype, pattern = cate)) +
geom_bar_pattern(
aes(pattern = cate),
stat = "identity",
position = position_dodge(width = 0.7),
width = 0.7,
color = "black", # Solid border
pattern_fill = "grey40", # Pattern color
pattern_angle = 45, # Angle of the dashed line
pattern_density = 0.05, # Spacing of the dashed line
pattern_spacing = 0.05 # Line spacing
) +
geom_text(
data = subset(data, P <= 0.05/3 & enrichment > 1),
aes(label = "*", y = enrichment + sdtimes * sd + 0.03, group = cate),
vjust = 0, color = "#F94144", size = 10, position = position_dodge(0.7)
) +
scale_pattern_manual(values = c("none", "stripe")) +
geom_errorbar(aes(ymin = enrichment - sdtimes*sd, ymax = enrichment + sdtimes*sd), width = 0.2, position = position_dodge(0.7), linewidth = 1) +
geom_hline(yintercept = 1, linetype = "dashed", color = "grey20", linewidth = 1) +
# scale_fill_brewer(palette = "Set2") +
scale_fill_manual(values = colors) +
labs(
title = "",
x = "",
y = "Excess Overlap"
) +
ylim(c(0,4.5)) +
theme_minimal(base_size = 15) +
theme(
plot.title = element_text(size = 0, face = "bold", hjust = 0.5),
axis.title.x = element_text(size = 0),
axis.title.y = element_text(size = 24),
axis.text.y = element_text(size = 20, margin = margin(r = 0), angle = 90, hjust = 0.5, vjust = 0),
axis.text.x = element_text(size = 20, margin = margin(t = 0)),
legend.position = "inside",
legend.justification = c(0.1, 0.9),
legend.title = element_text(size = 0),
legend.text = element_text(size = 20),
legend.spacing.y = unit(1.5, "cm"),
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5),
panel.grid.major = element_blank(), # Remove major grid lines
panel.grid.minor = element_blank(), # Remove minor grid lines
) +
guides(fill = "none",
pattern = guide_legend(
override.aes = list(
fill = c("white", "lightgrey"), # Adjust background color for the legend
color = "black" # Keep border color
)
)
)