Figure S10. Extended disease heritability analyses of variant-level functional annotations derived from ColocBoost.#
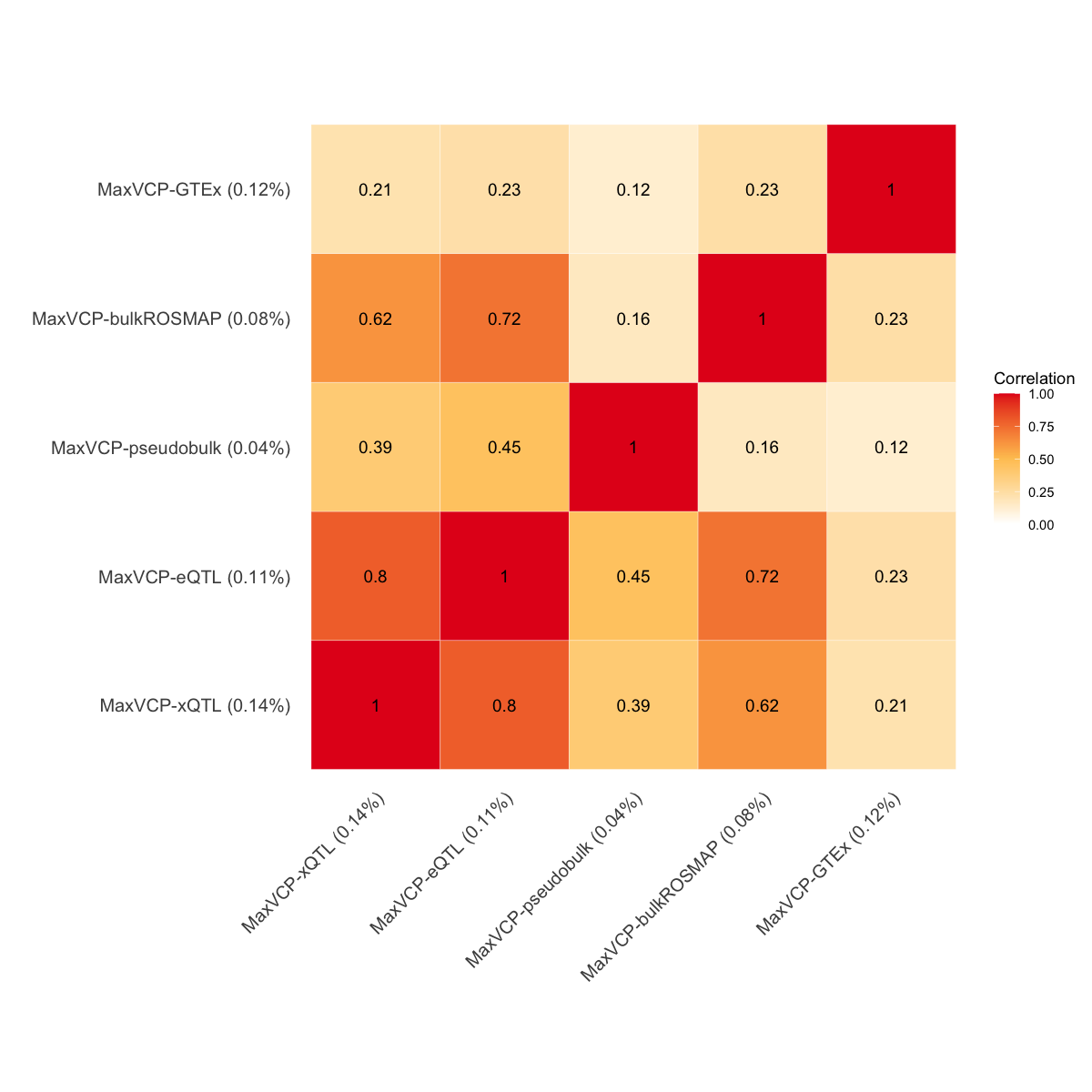
S10a. Correlation matrix of five functional annotations based on the MaxVCP scores from ColocBoost.
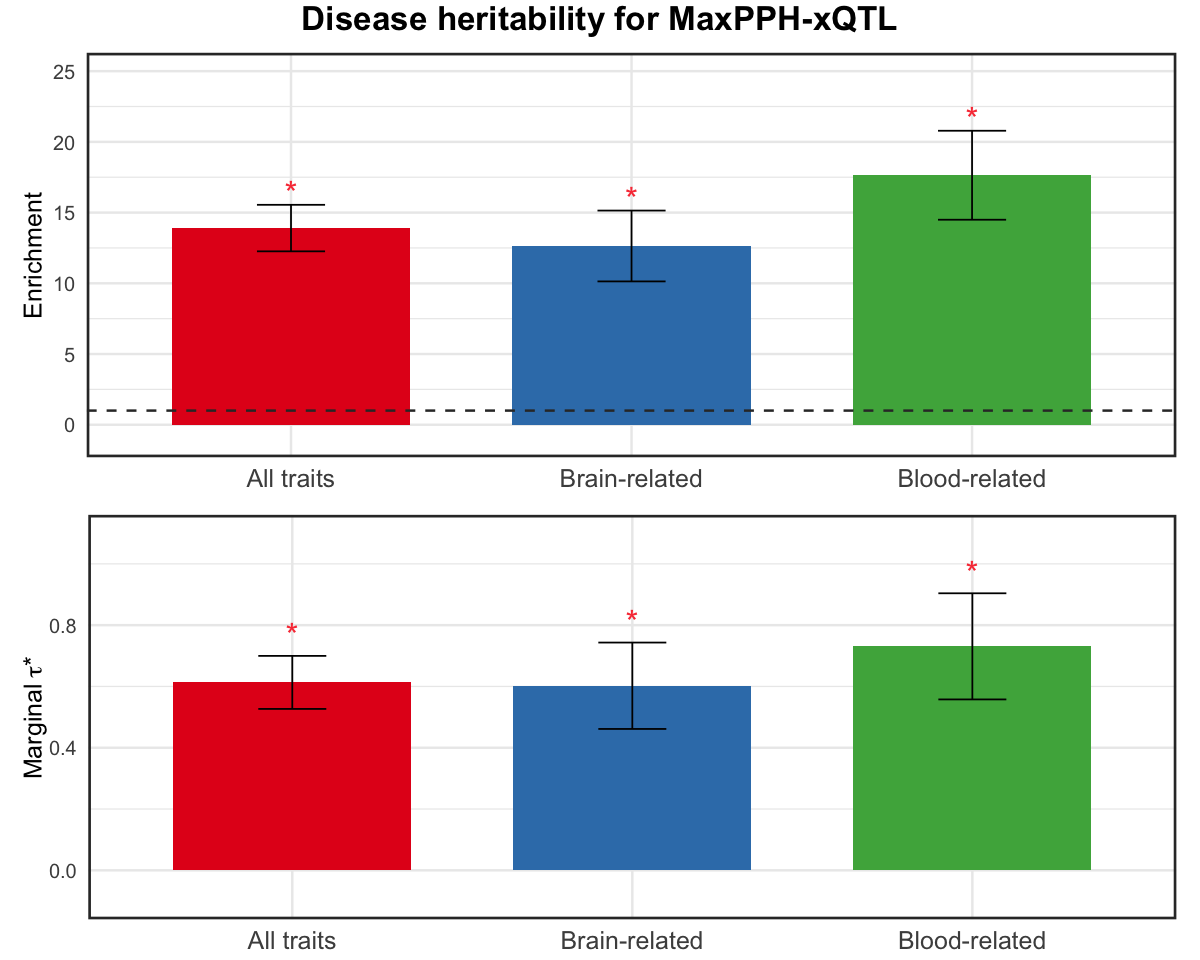
S10b. Heritability enrichment and standardized effect sizes, meta-analyzed across all, brain-related and blood-related traits, conditional on 97 baseline-LD v2.2 annotations of the 5 versions of xQTL MaxPPH derived from HyPrColoc.
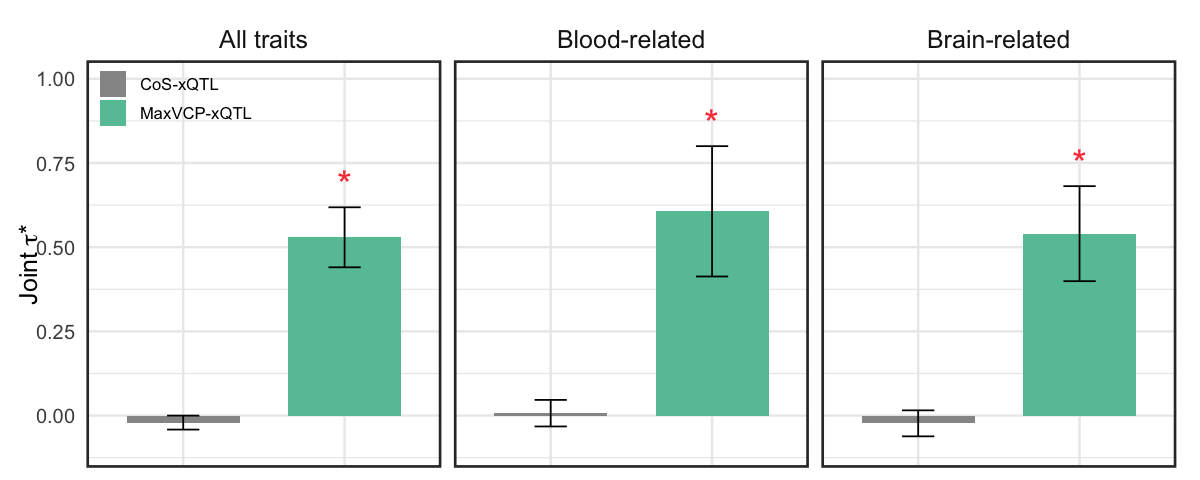
S10c. Standardized effect sizes, conditional on the 97 baseline-LD v2.2 annotations + MaxVCP-xQTL and CoS‐xQTL annotations (joint \(\tau^*\)).
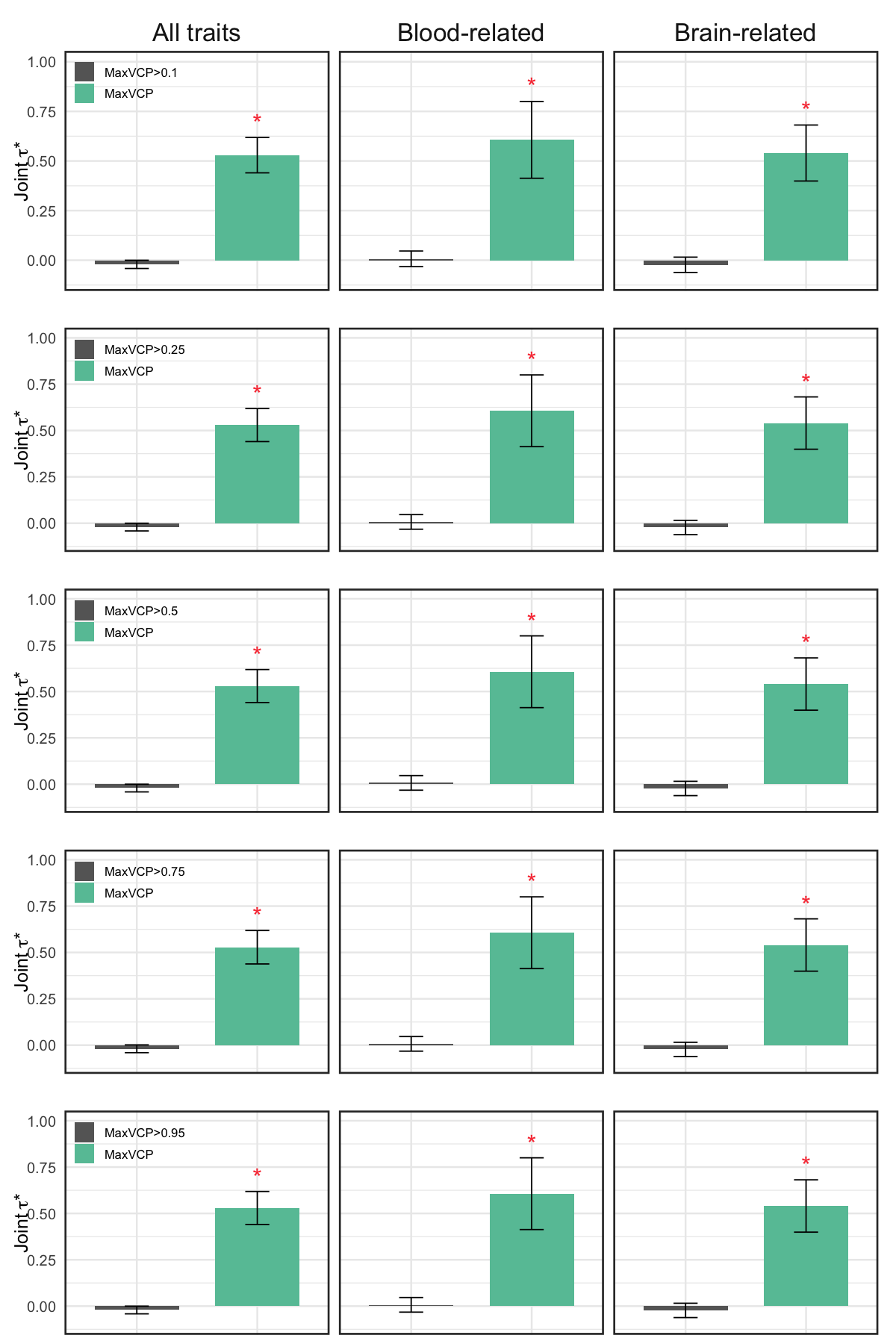
S10d. Standardized effect sizes, conditional on the 97 baseline-LD v2.2 annotations + MaxVCP-xQTL and binarized MaxVCP-xQTL scores at different binarization thresholds (joint \(\tau^*\)).
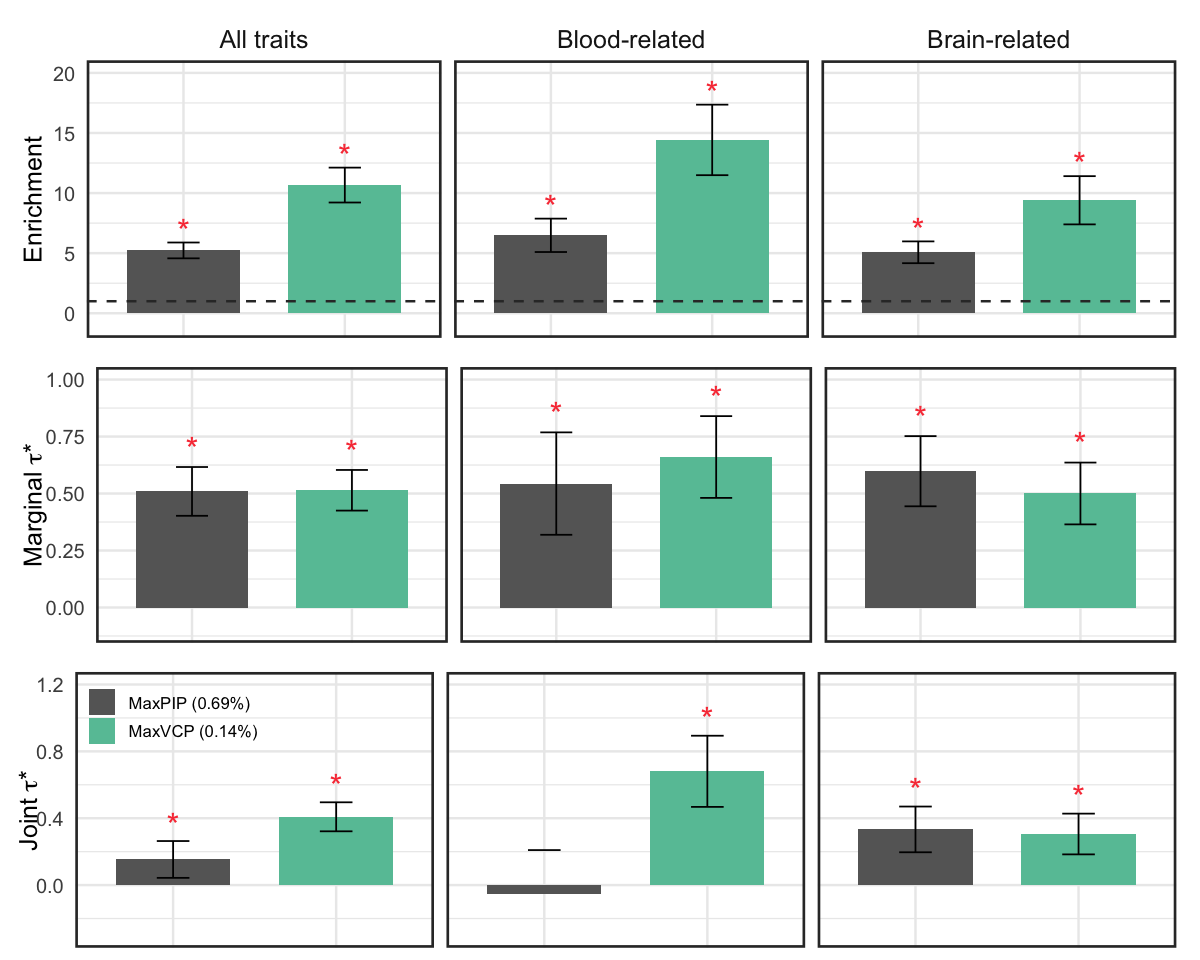
S10e. Heritability enrichment and standardized effect sizes (marginal \(\tau^*\) and joint \(\tau^*\)) of MaxVCP-xQTL against MaxPIP annotation from SuSiE fine-mapping analysis.
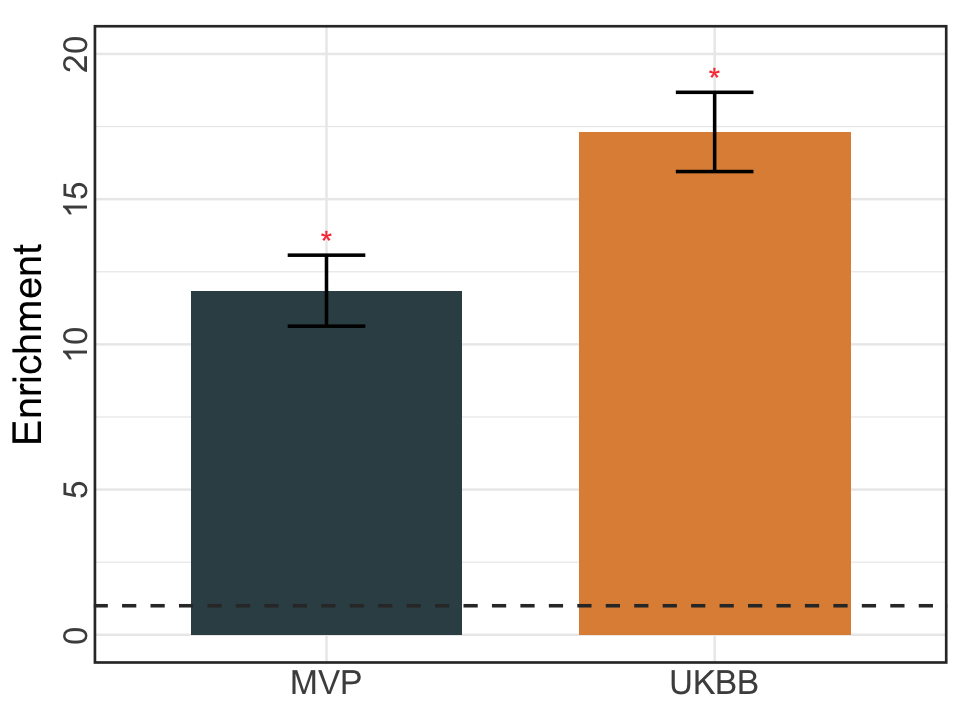
S10f. EOO analysis of MaxVCP-xQTL in the disease fine-mapped variants (PIP>0.95) from 94 UK Biobank traits and 930 Million Veteran Program (MVP) traits.
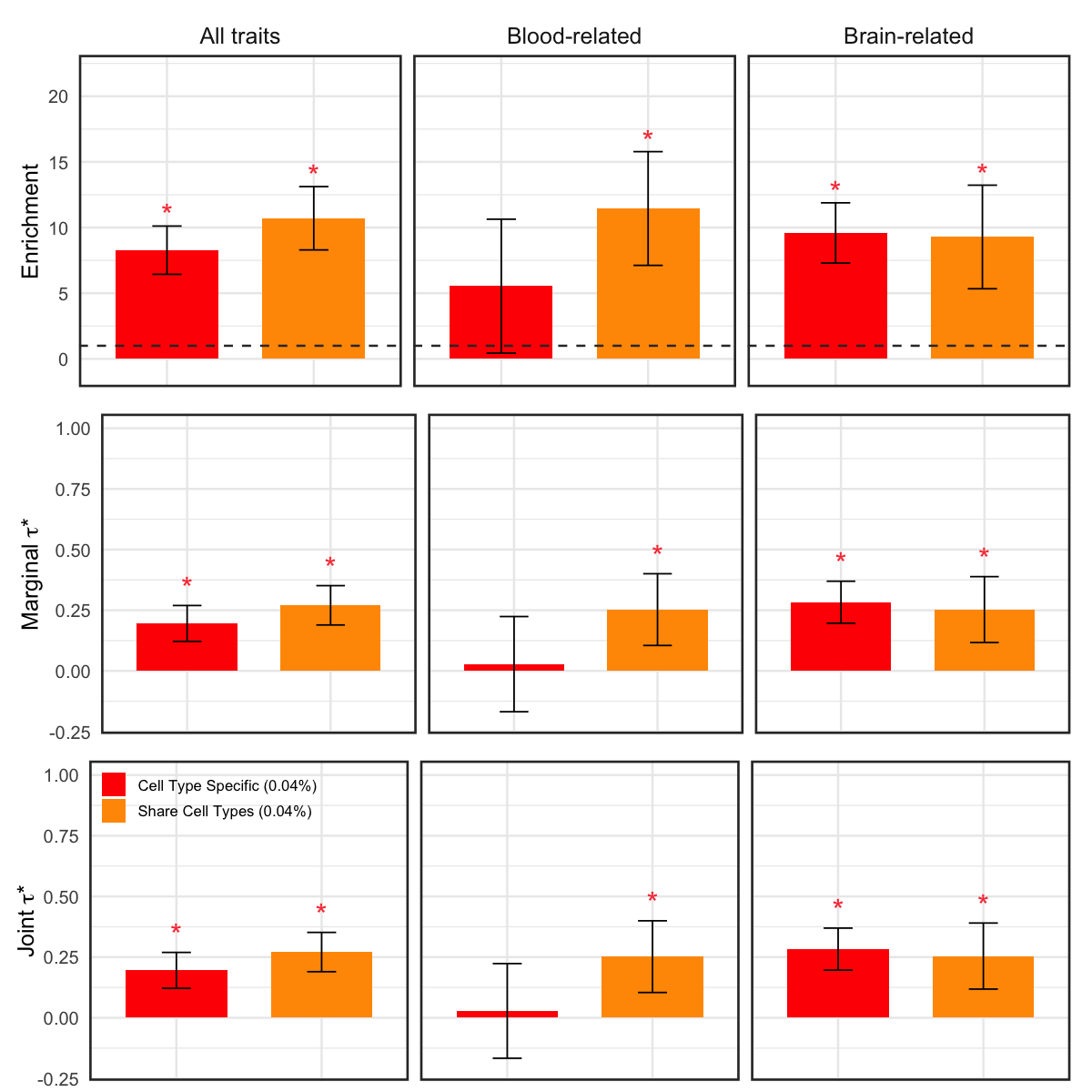
S10g. Heritability enrichment and standardized effect sizes (marginal \(\tau^*\) and joint \(\tau^*\)) of MaxVCP-xQTL annotations for cell‐type‐specific variants and cell‐type‐shared variants. Error bars denote 95% confidence intervals.
Asterisk denotes FDR<5% is computed by Bonferroni correction of the p-values for the number of annotations tested in the S-LDSC analysis.
Figure S10a#
Correlation matrix of five functional annotations based on the MaxVCP scores from ColocBoost.
correlation <- readRDS("Figure_S10a.rds")
library(ggplot2)
library(reshape2)
corr_melt <- melt(correlation)
p1 <- ggplot(corr_melt, aes(Var1, Var2, fill = value)) +
geom_tile(color = "white") +
scale_fill_gradient2(low = "white", high = "#E41C1C", mid = "#FEC661",
midpoint = 0.5, limit = c(0, 1), space = "Lab",
name="Correlation") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 12, hjust = 1),
axis.text.y = element_text(size = 12),
panel.grid = element_blank()) +
coord_fixed() +
geom_text(aes(Var1, Var2, label = round(value, 2)), color = "black", size = 4) +
labs(title = "", x = "", y = "")
Figure S10b#
Heritability enrichment and standardized effect sizes, meta-analyzed across all, brain-related and blood-related traits, conditional on 97 baseline-LD v2.2 annotations of the 5 versions of xQTL MaxPPH derived from HyPrColoc.
create_plot <- function(context){
if (context == "marginal tau*") {
y.show <- expression(paste("Marginal ", tau, "*"))
} else {
y.show = context
}
p <- ggplot(data[data$Context == context, ], aes(x = Disease, y = Value, fill = Disease)) +
geom_bar(stat = "identity", position = position_dodge(), width = 0.7) +
geom_errorbar(aes(ymin = Value - sd*Error, ymax = Value + sd*Error), width = 0.2, position = position_dodge(0.7)) +
scale_fill_brewer(palette = "Set1") +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "",
y = y.show,
fill = "Annotation"
) +
ylim(ylim_values[[context]])
if (context == "marginal tau*") {
p <- p + geom_text(data = subset(data, Context == "marginal tau*" & (P <= 0.05/6) & Value > 0), aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7)
} else {
p <- p + geom_hline(yintercept = 1, linetype = "dashed", color = "grey20", linewidth = 0.7) +
geom_text(data = subset(data, Context == "Enrichment" & (P <= 0.05/6) & Value > 0), aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7)
}
if (context == "Enrichment"){
p <- p + theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.5), "cm"),
strip.text.x = element_text(size = 15),
axis.text.x = element_text(size = 15),
legend.position = "none",
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
} else {
p <- p + theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.5), "cm"),
strip.text.x = element_blank(),
axis.text.x = element_text(size = 15),
axis.title.y = element_text(margin = margin(r = -2)),
legend.position = "none",
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
}
return(p)
}
data <- readRDS("Figure_S10b.rds")
library(ggplot2)
library(ggforce)
library(ggsci)
sd <- 1.96
ylim_values <- list(
"marginal tau*" = c(-0.1, 1.1),
"Enrichment" = c(-1, 25)
)
plot_single_tau <- create_plot("marginal tau*")
plot_enrichment <- create_plot("Enrichment")
library(ggpubr)
pp = ggarrange(plot_enrichment, plot_single_tau,
ncol = 1, nrow = 2,
heights = c(0.5,0.5))
Figure S10c#
Standardized effect sizes, conditional on the 97 baseline-LD v2.2 annotations + MaxVCP-xQTL and CoS‐xQTL annotations (joint \(\tau^*\)).
data <- readRDS("Figure_S10c.rds")
color <- c("#BDBDBD", "#969696", "#fc8d62", "#8da0cb", "#66c2a5")
ylim_values <- list(
"joint tau*" = c(-0.1, 1)
)
context <- "joint tau*"
y.show <- expression(paste("Joint ", tau, "*"))
p <- ggplot(data[data$Anno == "baseline LD", ], aes(x = Annotation, y = Value, fill = Annotation)) +
geom_bar(stat = "identity", position = position_dodge(), width = 0.7) +
geom_errorbar(aes(ymin = Value - sd*Error, ymax = Value + sd*Error), width = 0.2, position = position_dodge(0.7)) +
facet_wrap(~ Disease) +
scale_fill_manual(values = c("CoS-xQTL" = color[2],
"MaxVCP-xQTL" = color[5])) +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "",
y = y.show,
fill = "Annotation"
) +
ylim(ylim_values[[context]]) +
geom_text(data = subset(data, Anno == "baseline LD" & (P <= 0.05/5) & Value > 0),
aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 8) +
theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.4), "cm"),
strip.text.x = element_text(size = 15),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -4)),
legend.position = "inside",
legend.justification = c(0, 1.05),
legend.title = element_text(size = 0),
legend.text = element_text(size = 10),
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
Figure S10d#
Standardized effect sizes, conditional on the 97 baseline-LD v2.2 annotations + MaxVCP-xQTL and binarized MaxVCP-xQTL scores at different binarization thresholds (joint \(\tau^*\)).
sd <- 1.96
color <- c("#BDBDBD", "grey40", "#fc8d62", "#8da0cb", "#66c2a5")
ylim_values <- list(
"joint tau*" = c(-0.1, 1)
)
context <- "joint tau*"
y.show <- expression(paste("Joint ", tau, "*"))
data <- readRDS("Figure_S10d_05.rds")
p05 <- ggplot(data[data$Anno == "baseline LD", ], aes(x = Annotation, y = Value, fill = Annotation)) +
geom_bar(stat = "identity", position = position_dodge(), width = 0.7) +
geom_errorbar(aes(ymin = Value - sd*Error, ymax = Value + sd*Error), width = 0.2, position = position_dodge(0.7)) +
facet_wrap(~ Disease) +
scale_fill_manual(values = c("MaxVCP>0.5" = color[2],
"MaxVCP" = color[5])) +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "",
y = y.show,
fill = "Annotation"
) +
ylim(ylim_values[[context]]) +
geom_text(data = subset(data, Anno == "baseline LD" & (P <= 0.05/5) & Value > 0),
aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7) +
theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.4), "cm"),
strip.text.x = element_text(size = 0),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -4)),
legend.position = "inside",
legend.justification = c(0, 1.05),
legend.title = element_text(size = 0),
legend.text = element_text(size = 10),
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
data <- readRDS("Figure_S10d_025.rds")
p025 <- ggplot(data[data$Anno == "baseline LD", ], aes(x = Annotation, y = Value, fill = Annotation)) +
geom_bar(stat = "identity", position = position_dodge(), width = 0.7) +
geom_errorbar(aes(ymin = Value - sd*Error, ymax = Value + sd*Error), width = 0.2, position = position_dodge(0.7)) +
facet_wrap(~ Disease) +
scale_fill_manual(values = c("MaxVCP>0.25" = color[2],
"MaxVCP" = color[5])) +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "",
y = y.show,
fill = "Annotation"
) +
ylim(ylim_values[[context]]) +
geom_text(data = subset(data, Anno == "baseline LD" & (P <= 0.05/5) & Value > 0),
aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7) +
theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.4), "cm"),
strip.text.x = element_text(size = 0),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -4)),
legend.position = "inside",
legend.justification = c(0, 1.05),
legend.title = element_text(size = 0),
legend.text = element_text(size = 10),
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
data <- readRDS("Figure_S10d_075.rds")
p075 <- ggplot(data[data$Anno == "baseline LD", ], aes(x = Annotation, y = Value, fill = Annotation)) +
geom_bar(stat = "identity", position = position_dodge(), width = 0.7) +
geom_errorbar(aes(ymin = Value - sd*Error, ymax = Value + sd*Error), width = 0.2, position = position_dodge(0.7)) +
facet_wrap(~ Disease) +
scale_fill_manual(values = c("MaxVCP>0.75" = color[2],
"MaxVCP" = color[5])) +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "",
y = y.show,
fill = "Annotation"
) +
ylim(ylim_values[[context]]) +
geom_text(data = subset(data, Anno == "baseline LD" & (P <= 0.05/5) & Value > 0),
aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7) +
theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.4), "cm"),
strip.text.x = element_text(size = 0),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -4)),
legend.position = "inside",
legend.justification = c(0, 1.05),
legend.title = element_text(size = 0),
legend.text = element_text(size = 10),
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
data <- readRDS("Figure_S10d_01.rds")
p01 <- ggplot(data[data$Anno == "baseline LD", ], aes(x = Annotation, y = Value, fill = Annotation)) +
geom_bar(stat = "identity", position = position_dodge(), width = 0.7) +
geom_errorbar(aes(ymin = Value - sd*Error, ymax = Value + sd*Error), width = 0.2, position = position_dodge(0.7)) +
facet_wrap(~ Disease) +
scale_fill_manual(values = c("MaxVCP>0.1" = color[2],
"MaxVCP" = color[5])) +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "",
y = y.show,
fill = "Annotation"
) +
ylim(ylim_values[[context]]) +
geom_text(data = subset(data, Anno == "baseline LD" & (P <= 0.05/5) & Value > 0),
aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7) +
theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.4), "cm"),
strip.text.x = element_text(size = 20),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -4)),
legend.position = "inside",
legend.justification = c(0, 1.05),
legend.title = element_text(size = 0),
legend.text = element_text(size = 10),
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
data <- readRDS("Figure_S10d_095.rds")
p095 <- ggplot(data[data$Anno == "baseline LD", ], aes(x = Annotation, y = Value, fill = Annotation)) +
geom_bar(stat = "identity", position = position_dodge(), width = 0.7) +
geom_errorbar(aes(ymin = Value - sd*Error, ymax = Value + sd*Error), width = 0.2, position = position_dodge(0.7)) +
facet_wrap(~ Disease) +
scale_fill_manual(values = c("MaxVCP>0.95" = color[2],
"MaxVCP" = color[5])) +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "",
y = y.show,
fill = "Annotation"
) +
ylim(ylim_values[[context]]) +
geom_text(data = subset(data, Anno == "baseline LD" & (P <= 0.05/5) & Value > 0),
aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7) +
theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.4), "cm"),
strip.text.x = element_text(size = 0),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -4)),
legend.position = "inside",
legend.justification = c(0, 1.05),
legend.title = element_text(size = 0),
legend.text = element_text(size = 10),
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
Figure S10e#
Heritability enrichment and standardized effect sizes (marginal \(\tau^*\) and joint \(\tau^*\)) of MaxVCP-xQTL against MaxPIP annotation from SuSiE fine-mapping analysis.
data <- readRDS("Figure_S10e.rds")
ylim_values <- list(
"marginal tau*" = c(-0.1, 1),
"Enrichment" = c(-1, 20),
"joint tau*" = c(-0.3, 1.2)
)
create_plot <- function(context){
if (context == "marginal tau*") {
y.show <- expression(paste("Marginal ", tau, "*"))
} else if (context == "joint tau*") {
y.show <- expression(paste("Joint ", tau, "*"))
} else {
y.show = context
}
p <- ggplot(data[data$Context == context, ], aes(x = Annotation, y = Value, fill = Annotation)) +
geom_bar(stat = "identity", position = position_dodge(), width = 0.7) +
geom_errorbar(aes(ymin = Value - sd*Error, ymax = Value + sd*Error), width = 0.2, position = position_dodge(0.7)) +
facet_wrap(~ Disease) +
scale_fill_manual(values = c("MaxPIP (0.69%)" = color[2],
"MaxVCP (0.14%)" = color[5])) +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "",
y = y.show,
fill = "Annotation"
) +
ylim(ylim_values[[context]])
if (context == "marginal tau*") {
p <- p + geom_text(data = subset(data, Context == "marginal tau*" & (P <= 0.05/2) & Value > 0), aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7)
} else if (context == "joint tau*") {
p <- p + geom_text(data = subset(data, Context == "joint tau*" & (P <= 0.05/2) & Value > 0), aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7)
} else {
p <- p + geom_hline(yintercept = 1, linetype = "dashed", color = "grey20", linewidth = 0.7) +
geom_text(data = subset(data, Context == "Enrichment" & (P <= 0.05/2) & Value > 0), aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7)
}
if (context == "joint tau*"){
p <- p + theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.4), "cm"),
strip.text.x = element_blank(),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -4)),
legend.position = "inside",
legend.justification = c(0, 1.05),
legend.title = element_text(size = 0),
legend.text = element_text(size = 10),
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
} else if (context == "Enrichment"){
p <- p + theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.5), "cm"),
strip.text.x = element_text(size = 15),
axis.text.x = element_blank(),
legend.position = "none",
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
} else {
p <- p + theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.5), "cm"),
strip.text.x = element_blank(),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -4)),
legend.position = "none",
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
}
return(p)
}
plot_single_tau <- create_plot("marginal tau*")
plot_enrichment <- create_plot("Enrichment")
plot_joint_tau <- create_plot("joint tau*")
pp = ggarrange(plot_enrichment, plot_single_tau, plot_joint_tau,
ncol = 1, nrow = 3,
heights = c(0.31, 0.27, 0.27))
Figure S10f#
EOO analysis of MaxVCP-xQTL in the disease fine-mapped variants (PIP>0.95) from 94 UK Biobank traits and 930 Million Veteran Program (MVP) traits.
data <- readRDS("Figure_S10f.rds")
sdtimes <- 1.96
library(ggplot2)
library(ggsci)
p <- ggplot(data, aes(x = celltype, y = enrichment, fill = celltype)) +
geom_bar(stat = "identity", width = 0.7, position = position_dodge(width = 0.9)) +
geom_errorbar(aes(ymin = enrichment - sdtimes*sd, ymax = enrichment + sdtimes*sd), width = 0.2, position = position_dodge(0.7), linewidth = 1) +
geom_text(aes(label = "*", y = enrichment + sd * sdtimes + 0.03), vjust = 0, color = "#F94144", size = 7) +
geom_hline(yintercept = 1, linetype = "dashed", color = "grey20", linewidth = 1) +
# scale_fill_brewer(palette = "Spectral", direction = -1) +
scale_fill_jama() +
labs(
title = "",
x = "",
y = "Enrichment"
) +
ylim(c(0,20)) +
theme_minimal(base_size = 15) +
theme(
plot.title = element_text(size = 0, face = "bold", hjust = 0.5),
axis.title.x = element_text(size = 0),
axis.title.y = element_text(size = 24),
axis.text.y = element_text(size = 20, margin = margin(r = 0), angle = 90, hjust = 0.5, vjust = 0),
axis.text.x = element_text(size = 20, margin = margin(t = 0)),
legend.position = "none",
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
Figure S10g#
Heritability enrichment and standardized effect sizes (marginal \(\tau^*\) and joint \(\tau^*\)) of MaxVCP-xQTL annotations for cell‐type‐specific variants and cell‐type‐shared variants. Error bars denote 95% confidence intervals.
data <- readRDS("Figure_S10g.rds")
sd <- 1.96
# color <- c(pal_npg()(10), pal_d3()(10))
color <- c("#BDBDBD", "#969696", "#fc8d62", "#8da0cb", "#66c2a5")
ylim_values <- list(
"marginal tau*" = c(-0.2, 1),
"Enrichment" = c(-1, 22),
"joint tau*" = c(-0.2, 1)
)
create_plot <- function(context){
if (context == "marginal tau*") {
y.show <- expression(paste("Marginal ", tau, "*"))
} else if (context == "joint tau*") {
y.show <- expression(paste("Joint ", tau, "*"))
} else {
y.show = context
}
p <- ggplot(data[data$Context == context, ], aes(x = Annotation, y = Value, fill = Annotation)) +
geom_bar(stat = "identity", position = position_dodge(), width = 0.7) +
geom_errorbar(aes(ymin = Value - sd*Error, ymax = Value + sd*Error), width = 0.2, position = position_dodge(0.7)) +
facet_wrap(~ Disease) +
scale_fill_ucscgb() +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "",
y = y.show,
fill = "Annotation"
) +
ylim(ylim_values[[context]])
if (context == "marginal tau*") {
p <- p + geom_text(data = subset(data, Context == "marginal tau*" & (P <= 0.05/2) & Value > 0), aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7)
} else if (context == "joint tau*") {
p <- p + geom_text(data = subset(data, Context == "joint tau*" & (P <= 0.05/2) & Value > 0), aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7)
} else {
p <- p + geom_hline(yintercept = 1, linetype = "dashed", color = "grey20", linewidth = 0.7) +
geom_text(data = subset(data, Context == "Enrichment" & (P <= 0.05/2) & Value > 0), aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7)
}
if (context == "joint tau*"){
p <- p + theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.4), "cm"),
strip.text.x = element_blank(),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -4)),
legend.position = "inside",
legend.justification = c(0, 1.05),
legend.title = element_text(size = 0),
legend.text = element_text(size = 10),
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
} else if (context == "Enrichment"){
p <- p + theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.5), "cm"),
strip.text.x = element_text(size = 15),
axis.text.x = element_blank(),
legend.position = "none",
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
} else {
p <- p + theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.5), "cm"),
strip.text.x = element_blank(),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -2)),
legend.position = "none",
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
}
return(p)
}
plot_single_tau <- create_plot("marginal tau*")
plot_enrichment <- create_plot("Enrichment")
plot_joint_tau <- create_plot("joint tau*")
library(ggpubr)
pp = ggarrange(plot_enrichment, plot_single_tau, plot_joint_tau,
ncol = 1, nrow = 3,
heights = c(0.31, 0.27, 0.27))