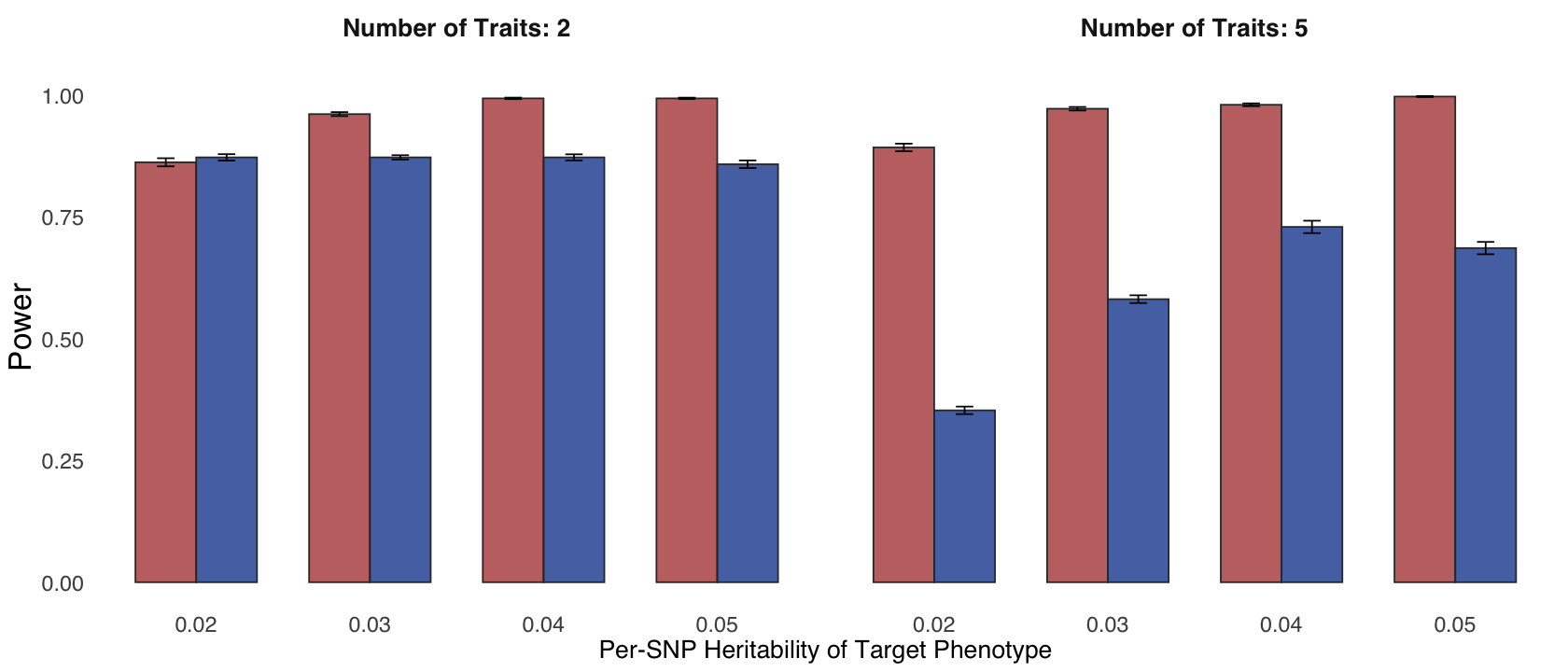
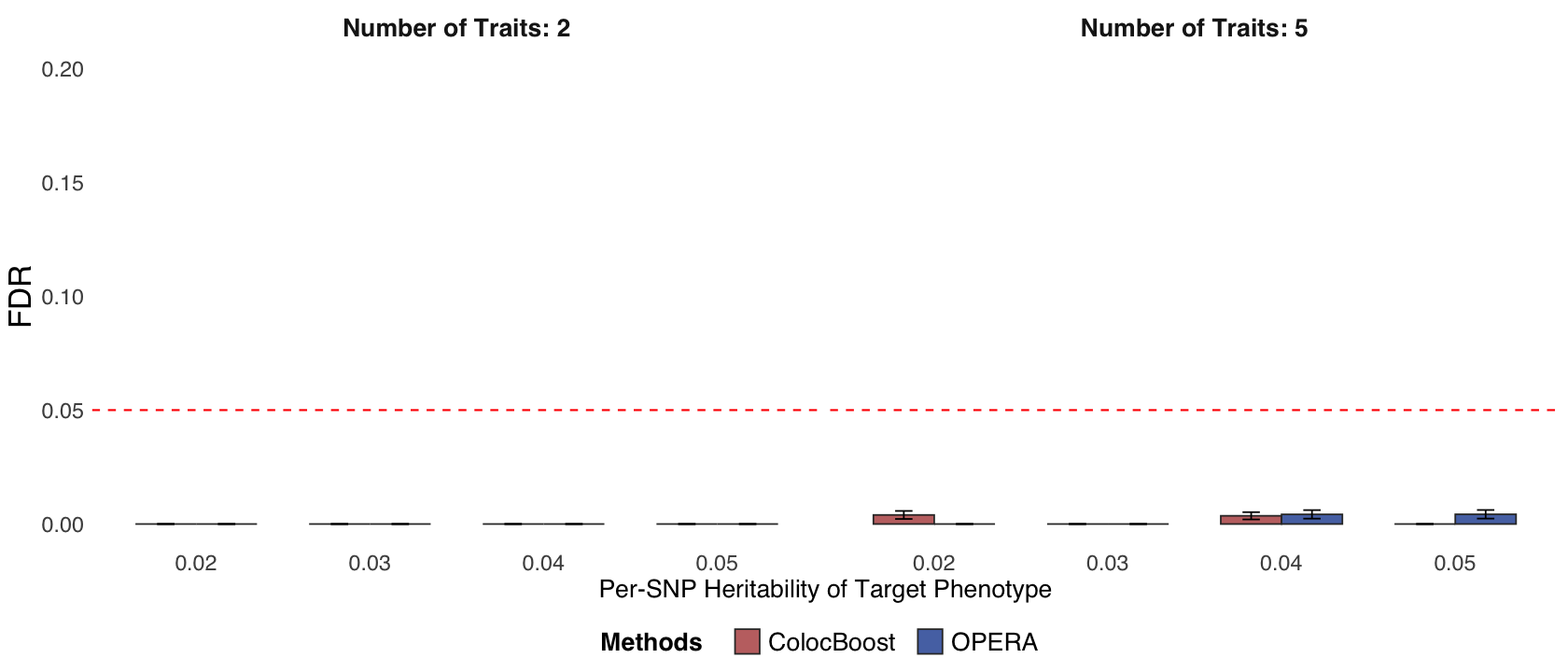
Figure 2e. Statistical power and False Discovery Rate (FDR) comparisons.#
Statistical power and FDR comparison of ColocBoost with OPERA for GWAS colocalization, evaluated at the gene level (Methods).
library(tidyverse)
library(ggpattern)
library(ggpubr)
library(cowplot)
result <- readRDS("data/Figure_2e.rds")
result$method <- factor(result$method, levels = c("ColocBoost", "OPERA"))
colors_man <- c("#B24745FF", "#1d4a9e")
Statistical power#
p1 <- result %>%
ggplot(aes(x = as.factor(phi), y = power, fill = method)) +
facet_wrap( ~ trait, labeller = labeller(trait = function(x) paste("Number of Traits:", x)), scales = "free_x", nrow = 1) +
geom_col(position = "dodge", width = 0.7, colour = "grey20", alpha = 0.8) +
theme_minimal() +
ylim(0, 1.05) + # Ensures x-axis ticks are integers
geom_errorbar(aes(ymin = power - 1.96*power_SD/ sqrt(20) ,
ymax = power + 1.96*power_SD/ sqrt(20) ),
width = .2, position = position_dodge(width = 0.7)) +
scale_fill_manual(values = colors_man, name = "Methods") +
# guides(fill = guide_legend(title = "Methods")) +
labs(x = "Per-SNP Heritability of Target Phenotype", y = "Power", color = "Methods") +
theme(legend.position = "none",
axis.title.x = element_text(size = 16),
axis.title.y = element_text(size = 20),
axis.text.x = element_text(size = 14),
axis.text.y = element_text(size = 14),
strip.text = element_text(size = 16, face = "bold"),
legend.title = element_text(size = 16, face = "bold", margin = margin(l = 0, r= 20)), # Change legend title font size and style
legend.text = element_text(size = 16, margin = margin(l = 5, r= 8)),
text=element_text(size=16, family="sans")) + theme(
panel.grid.major = element_blank(), # Remove major grid lines
panel.grid.minor = element_blank(), # Remove minor grid lines
# axis.line = element_line(color = "black") # Keep the axis lines
)
FDR#
p2 <- result %>%
ggplot(aes(x = as.factor(phi), y = FDR, fill = method)) +
geom_col(position = "dodge", width = 0.7, colour = "grey20", alpha = 0.8) +
theme_minimal() +
facet_wrap( ~ trait, labeller = labeller(trait = function(x) paste("Number of Traits:", x)), scales = "free_x", nrow = 1) +
ylim(0, 0.2) + # Ensures x-axis ticks are integers
geom_errorbar(aes(ymin = FDR - 1.96 * FDR_SD / sqrt(20),
ymax = FDR + 1.96 *FDR_SD / sqrt(20)),
width = .2, position = position_dodge(width = 0.7)) +
scale_fill_manual(values = colors_man, name = "Methods") +
labs(x = "Per-SNP Heritability of Target Phenotype", y = "FDR", color = "Methods") +
geom_hline(yintercept = 0.05, linetype = "dashed", color = "red", linewidth = 0.6) +
theme(legend.position = "bottom",
axis.title.x = element_text(size = 16),
axis.title.y = element_text(size = 20),
axis.text.x = element_text(size = 14),
axis.text.y = element_text(size = 14),
strip.text = element_text(size = 16, face = "bold"),
legend.title = element_text(size = 16, face = "bold", margin = margin(l = 0, r= 20)), # Change legend title font size and style
legend.text = element_text(size = 16, margin = margin(l = 5, r= 8)),
text=element_text(size=16, family="sans")) + theme(
panel.grid.major = element_blank(), # Remove major grid lines
panel.grid.minor = element_blank(), # Remove minor grid lines
# axis.line = element_line(color = "black") # Keep the axis lines
)