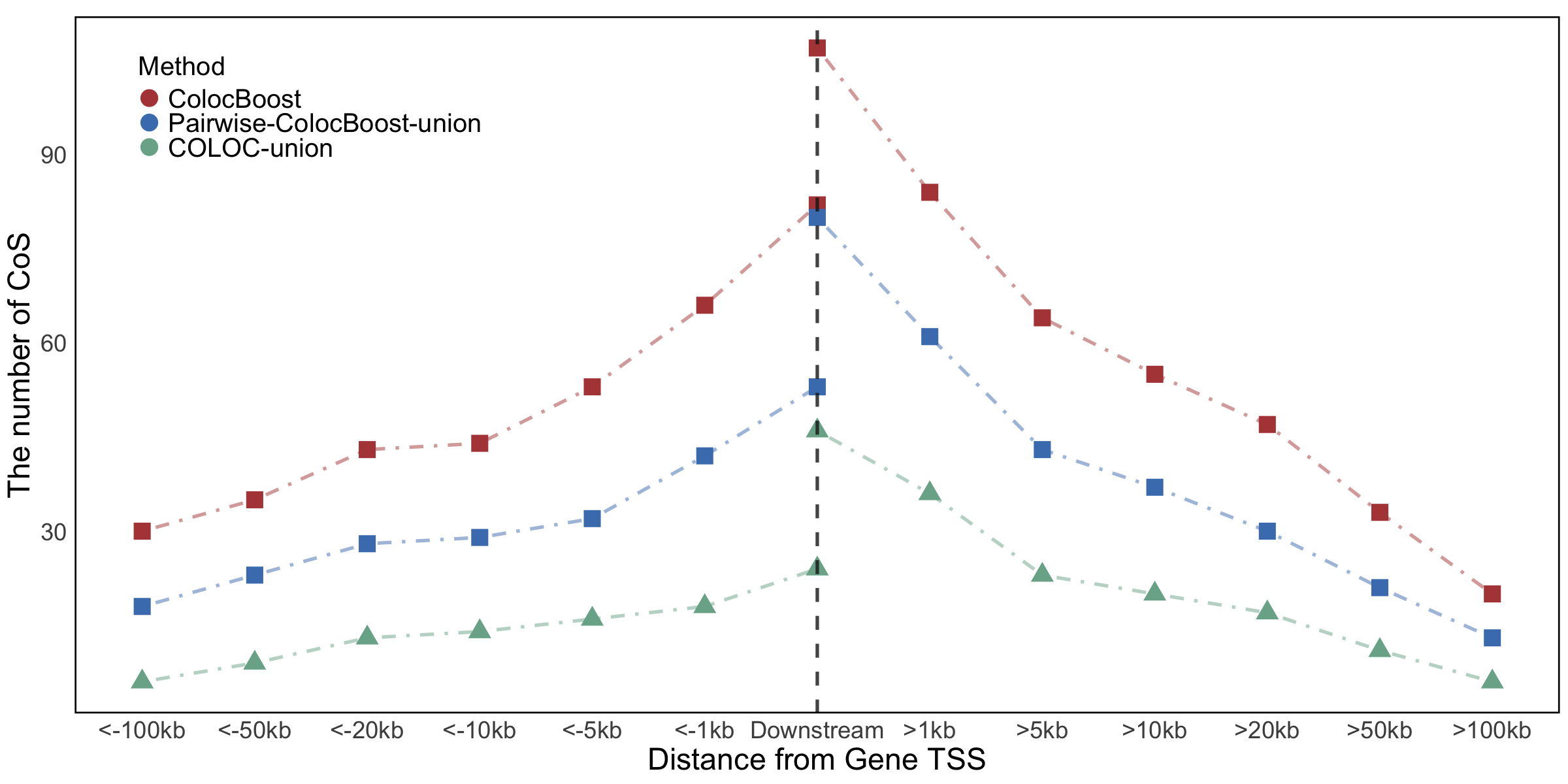
Figure 6c. Distribution of distances from the gene TSS for CoS.#
Distribution of distances from the gene TSS for CoS from AD–xQTL ColocBoost, COLOC-union, and the union of CoS identified by ColocBoost applied pairwise to one AD GWAS and each of xQTL traits (pairwise-ColocBoost-union).
data <- readRDS("data/Figure_6c.rds")
library(ggplot2)
p1 <- ggplot(data, aes(x = Dist, y = cumsum_count, color = group, shape = group)) +
geom_point(size = 7) +
geom_line(data = subset(data, cate == "Upstream"), aes(group = group), linewidth = 1.5, alpha = 0.5, linetype = 4) +
geom_line(data = subset(data, cate == "Downstream"), aes(group = group), linewidth = 1.5, alpha = 0.5, linetype = 4) +
geom_vline(xintercept = 7, color = "grey10", linetype = 2, linewidth = 1.5, alpha = 0.8) +
scale_shape_manual(values = c("COLOC-union" = 17, "ColocBoost" = 15, "Pairwise-ColocBoost-union" = 15)) +
guides(shape = "none") +
scale_color_manual(values = c("COLOC-union" = "#79AF97FF",
"Pairwise-ColocBoost-union" = "#4A7EBBFF",
"ColocBoost" = "#B24745FF")) +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "Distance from Gene TSS",
y = "The number of CoS",
color = "Method",
shape = "Category"
) +
theme(
plot.title = element_text(size = 0),
axis.title.x = element_text(size = 28),
axis.title.y = element_text(size = 28),
axis.text.x = element_text(size = 22), #, angle = 30, vjust = 0.5, hjust = 0.5),
axis.text.y = element_text(size = 22),
legend.title = element_text(size = 24),
legend.text = element_text(size = 24),
legend.position = "inside",
strip.text = element_text(size = 0, face = "bold"),
legend.justification = c(0.05, 0.95),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_rect(color = "black", fill = NA, linewidth = 1.5)
)