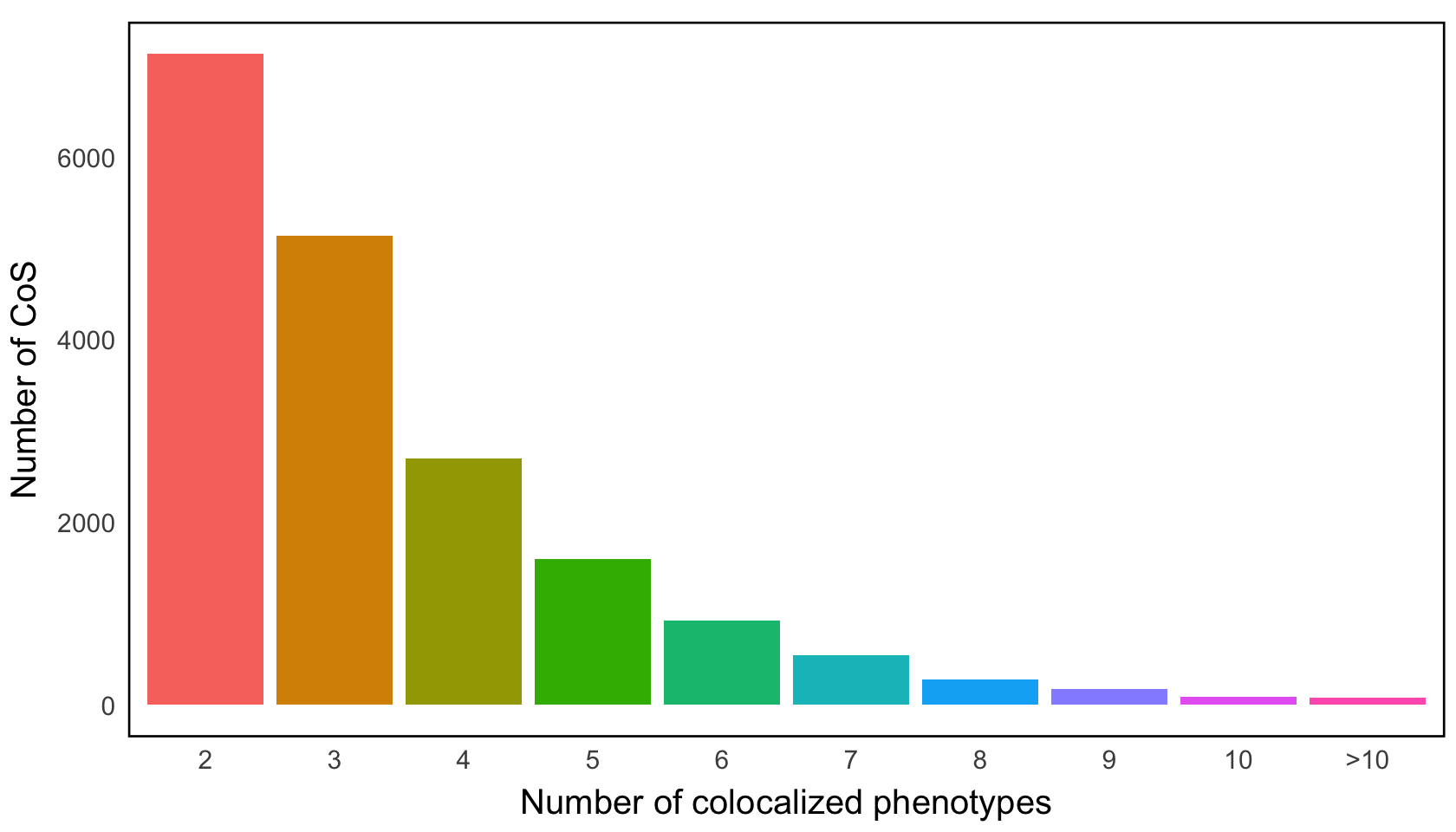
Figure 3b. Distribution of the number of 95% CoSs corresponding to the different numbers of colocalized traits per locus.#
Distribution of the number of 95% CoS corresponding to the different numbers of colocalized traits per locus; we omit from consideration 0.55% loci with more than 10 colocalized traits.
library(tidyverse)
library(ggpattern)
library(ggpubr)
library(cowplot)
res <- readRDS("data/xQTL_only_colocalization.rds")
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.4 ✔ tidyr 1.3.1
✔ purrr 1.0.4
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
Attaching package: ‘cowplot’
The following object is masked from ‘package:ggpubr’:
get_legend
The following object is masked from ‘package:lubridate’:
stamp
Organize input data#
coloc_pheno_numbers <- sapply(res$colocalized_phenotypes, function(cp){
length(strsplit(cp, "; ")[[1]])
})
table_numphen <- table(coloc_pheno_numbers)
data <- data.frame(num_phen = as.numeric(table_numphen),
categories = names(table_numphen))
data$proportion <- data$num_phen / sum(data$num_phen)
prop_10 <- sum(data$proportion[10:13])
num_10 <- sum(data$num_phen[10:13])
data <- data[1:9,]
data <- rbind(data, c(num_10, ">10", prop_10))
data <- within(data, {
categories <- factor(categories, levels = c(2,3,4,5,6,7,8,9,10,">10"))
num_phen <- as.numeric(num_phen)
proportion <- as.numeric(proportion)
})
Distribution plot#
library(ggsci)
color <- c(pal_npg()(10), pal_d3()(10))
p1 <- ggplot(data, aes(x = categories, y = num_phen, fill = categories)) +
geom_bar(stat = "identity") +
scale_color_manual(values = color) +
labs(
title = "",
x = "Number of colocalized phenotypes",
y = "Number of CoS"
) +
theme_minimal(base_size = 15) + # Use a minimal theme with a larger base font size
theme(
plot.title = element_text( size = 0 ),
axis.title.x = element_text( margin = margin(t = 10), size = 24), # Adjust x axis title margin
axis.title.y = element_text(margin = margin(r = 10), size = 24), # Adjust y axis title margin
axis.text.x = element_text(margin = margin(t = 5), size = 18), # Adjust x axis text margin
axis.text.y = element_text(margin = margin(r = 5), size = 18), # Adjust y axis text margin
legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_rect(color = "black", fill = NA, linewidth = 1.5)
)