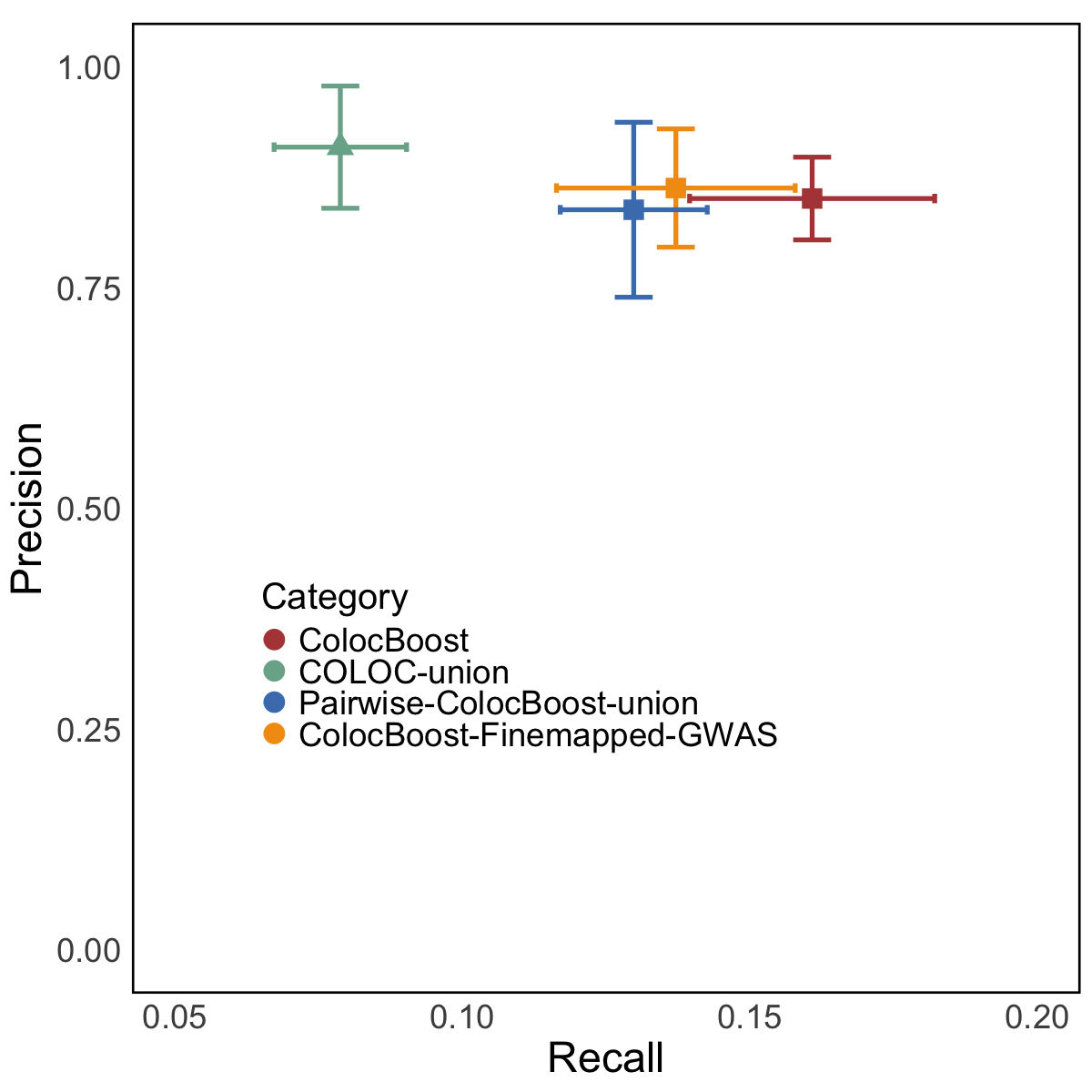
Figure 6d. Precision-recall analysis compared with ENCODE-rE2G links.#
Precision-recall analysis comparing CoS-gene links from AD-xQTL ColocBoost, COLOC-union, pairwise-ColocBoost-union, and a version limiting AD-xQTL ColocBoost to AD fine-mapped variants (ColocBoost-finemapped-GWAS) against enhancer-gene links predicted by ENCODE-rE2G across 354 biosamples. Error bars along both axis indicate 95% confidence intervals.
data <- readRDS("data/Figure_6d.rds")
library(ggplot2)
library(ggplot2)
sd <- 1.96
p1 <- ggplot(data, aes(x = recall, y = precision, color = method, shape = method)) +
geom_point(size = 6) +
scale_shape_manual(values = c("COLOC-union" = 17, "ColocBoost" = 15, "Pairwise-ColocBoost-union" = 15, "ColocBoost-Finemapped-GWAS" = 15)) +
guides(shape = "none") +
geom_errorbar(aes(ymin = precision - sd*precision_sd, ymax = precision + sd*precision_sd), linewidth = 1.5) +
geom_errorbarh(aes(xmin = recall - sd*recall_sd1, xmax = recall + sd*recall_sd1), linewidth = 1.5) +
scale_color_manual(values = c("COLOC-union" = "#79AF97FF",
"Pairwise-ColocBoost-union" = "#4A7EBBFF",
"ColocBoost" = "#B24745FF",
"ColocBoost-Finemapped-GWAS" = "#F39C12FF")) +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "Recall",
y = "Precision",
color = "Category",
shape = "Method"
) +
ylim(c(0, 1)) +
xlim(c(0.05, 0.2)) +
theme(
plot.title = element_text(size = 0),
axis.title.x = element_text(size = 28),
axis.title.y = element_text(size = 28),
axis.text.x = element_text(size = 22),
axis.text.y = element_text(size = 22),
legend.title = element_text(size = 24),
legend.text = element_text(size = 22),
legend.position = "inside",
legend.justification = c(0.3, 0.3),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_rect(color = "black", fill = NA, linewidth = 1.5)
)