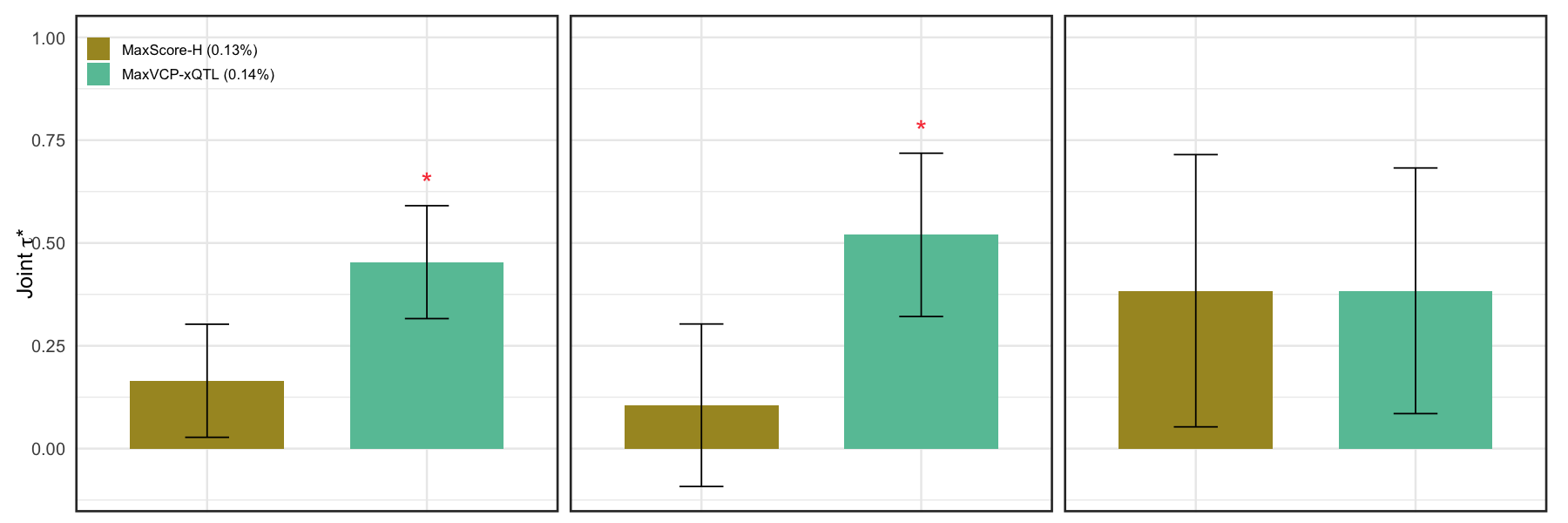
Figure 5d. Standardized effect sizes of the MaxVCP-xQTL and an analogous score based on the HyPrColoc method.#
Standardized effect sizes of the MaxVCP-xQTL and an analogous score based on the HyPrColoc method in a joint model involving other 97 baseline-LD v2.2 annotations. All results are meta-analyzed across 57 complex traits as well as subsets of 18 brain-related and 22 blood-related traits following from S-LDSC recommendations. The asterisks indicate statistical significance (Bonferroni adjusted p-value<0.05). Error bars indicate 95% confidence intervals.
library(ggplot2)
library(ggforce)
library(ggsci)
data <- readRDS("data/Figure_5d.rds")
sd <- 1.96
# color <- c(pal_npg()(10), pal_d3()(10))
color <- c("#a8952a", "#66c2a5")
ylim_values <- list(
"marginal tau*" = c(-0.1, 1),
"Enrichment" = c(-1, 22),
"joint tau*" = c(-0.1, 1)
)
create_plot <- function(context){
if (context == "marginal tau*") {
y.show <- expression(paste("Marginal ", tau, "*"))
} else if (context == "joint tau*") {
y.show <- expression(paste("Joint ", tau, "*"))
} else {
y.show = context
}
p <- ggplot(data[data$Context == context, ], aes(x = Annotation, y = Value, fill = Annotation)) +
geom_bar(stat = "identity", position = position_dodge(), width = 0.7) +
geom_errorbar(aes(ymin = Value - sd*Error, ymax = Value + sd*Error), width = 0.2, position = position_dodge(0.7)) +
facet_wrap(~ Disease) +
scale_fill_manual(values = c("MaxScore-H (0.13%)" = color[1],
"MaxVCP-xQTL (0.14%)" = color[2])) +
theme_minimal(base_size = 15) +
labs(
title = "",
x = "",
y = y.show,
fill = "Annotation"
) +
ylim(ylim_values[[context]])
if (context == "marginal tau*") {
p <- p + geom_text(data = subset(data, Context == "marginal tau*" & (P <= 0.05/6) & Value > 0), aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7)
} else if (context == "joint tau*") {
p <- p + geom_text(data = subset(data, Context == "joint tau*" & (P <= 0.05/6) & Value > 0), aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7)
} else {
p <- p + geom_hline(yintercept = 1, linetype = "dashed", color = "grey20", linewidth = 0.7) +
geom_text(data = subset(data, Context == "Enrichment" & (P <= 0.05/6) & Value > 0), aes(label = "*", y = Value + sd * Error + 0.03), vjust = 0, color = "#F94144", size = 7)
}
if (context == "joint tau*"){
p <- p + theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.4), "cm"),
strip.text.x = element_blank(),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -4)),
legend.position = "inside",
legend.justification = c(0, 1),
legend.title = element_text(size = 0),
legend.text = element_text(size = 10),
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
} else if (context == "Enrichment"){
p <- p + theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.5), "cm"),
strip.text.x = element_text(size = 15),
axis.text.x = element_blank(),
legend.position = "none",
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
} else {
p <- p + theme(
plot.margin = unit(c(-0.5, 0.5, -0.5, 0.5), "cm"),
strip.text.x = element_blank(),
axis.text.x = element_blank(),
axis.title.y = element_text(margin = margin(r = -2)),
legend.position = "none",
panel.border = element_rect(color = "grey20", fill = NA, linewidth = 1.5)
)
}
return(p)
}
plot_joint_tau <- create_plot("joint tau*")